How is transcriptional regulation of lipogenesis linked to diet and environment in C. elegans and mammals
SREBP (sterol regulatory element binding protein) transcription factors activate genes important for cholesterol metabolism, fatty acid (FA) and synthesis of phospholipids, in addition to ensuring production of additional important co-factors. We have found unexplored regulatory links between lipid metabolism and the one-carbon cycle (1CC), discovering that SREBP proteins in C. elegans and mammals control expression of 1CC genes. The 1CC provides methyl groups for PL biosynthesis and epigenetic regulation; regulation by SREBPs provides a novel layer of nutrition-dependent input to methylation-dependent processes. Alterations in 1C metabolism and SREBP function are associated with similar diseases, suggesting that co-regulation with lipid homeostasis may be a common impact point in metabolic-associated disorders such as obesity and fatty liver disease.
Most methylation reactions require SAMe (s-adenosyl methionine). Conversion of methionine to SAMe by MAT1A/MAT2A in humans, or the SAMS proteins in C. elegans, affects cellular processes from phosphatidylcholine (PC) biosynthesis to protein or DNA modification. We have found that SREBPs regulate sams-1 and MAT1A expression in both C. elegans and human cells. In C. elegans, sams-1 decrease or loss causes lipid accumulation reminiscent of hepatic steatosis occuring in MAT1A KO mice (Lu et al. PNAS 2001), suggesting models for lipid accumulation in sams-1 animals may be relevant to hepatic steatosis in mammals. Because the 1CC is implicated in multiple fatty liver models and metabolites such as folate or choline are administered as treatments for disease, it is crucial to understand the connections between the 1CC and lipid biosynthesis.
We are interested in how lipid homeostasis is affected by genetic or dietary changes in 1CC function and how SBP-1/SREBP affects cellular processes such as epigenetic modification by regulating the supply of methyl donors. These projects will include mechanistic studies in mammalian cell culture, in vivo studies in mouse liver as well as genetics screens for discovery in C. elegans. In our studies of SREBP regulation by SIRT1 (Walker, et al. 2010, Genes and Development) and of SREBP regulation of the 1CC (Walker, A., Jacobs, R. et al., Cell, 2011), this combination of models has allowed us to rapidly establish functional biological relationships between pathways in C. elegans, then determine relevance to mammalian physiology and precise molecular mechanisms in mouse knockout or human cell culture models. These types of experiments are important for determining mechanistic relationships between metabolic pathways and cellular function, and furthermore, will aid in our understanding about how these pathways contribute to human disorders such as metabolic syndrome.
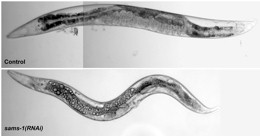
Figure. In C. elegans, SBP-1-Dependent Lipogenesis and Gene Expression Are Increased after sams-1(RNAi)
(A) RNAi knockdown of sams-1 revealed large refractile droplets in the intestine and body cavity by Nomarski optics. | 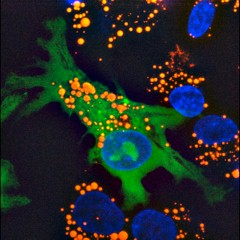 |